2. 临沂市肿瘤医院
相关研究显示,核内不均一核糖核蛋白A2/B1(HNRNPA2B1)是hnRNPs家族的一员,其通过识别并结合特定的RNA底物,参与转录、剪接、转运和翻译而调控下游基因表达,进而影响多个细胞生理过程[1-6]。HNRNPA2B1在多种恶性肿瘤组织和癌细胞系中高表达,与肿瘤生长、转移、血管生成和化疗耐药有关[7-14]。有研究显示,在非小细胞肺癌(NSCLC)中,HNRNPA2B1通过m6A依赖的方式调控miR-106b-5p[15-16]。众所周知,TP53是最常见的抑癌基因,其突变可见于35%~60%的肺腺癌中[17-19],且携带TP53突变的肺癌病人预后差[20];HNRNPA2B1可能通过促进miRNA成熟而影响肺腺癌病人预后[21]。但二者是否有相关性尚无报道。本研究通过数据库探究潜在受HNRNPA2B1调控的预后相关miRNAs,并构建TP53突变肺腺癌预后模型,探讨HNRNPA2B1-miRNAs在TP53突变肺腺癌病人预后中的作用及其机制。
1 材料与方法 1.1 公共数据获取通过癌症基因组图谱(TCGA)和基因表达数据库(GEO)官网下载59例正常对照样本和526例肺腺癌样本的基因突变、mRNA和miRNAs表达数据,并下载与之对应的病人临床信息用来进行预后分析。根据肺腺癌常见突变基因TP53将肺腺癌病人分为TP53突变组(TP53-mut组)和TP53野生型组(TP53-wt组);并将病人随机分为训练集和验证集,比例为7∶3。
1.2 基因集富集分析(GSEA)GSEA可以通过将预定义的基因集与特定表型进行比对从而找出与预定义基因集相关的表型。根据HNRNPA2B1表达量的高低进行分组,对本研究基因在肺腺癌中可能的作用机制进行探索。
1.3 变量筛选Spearman相关性分析以相关系数ρ>0且P<0.05为筛选条件,选出与HNRNPA2B1表达呈正相关的miRNAs。应用Kaplan-Meier预后分析选出在TP53突变肺腺癌病人中表达越高预后越差的miRNAs,将同时符合以上两个条件的miRNAs进一步筛选,最后纳入预后模型构建。
1.4 模型构建和评估利用HNRNPA2B1及其调控miRNAs进行预后模型构建,通过风险得分中位值将样本分为高风险、低风险两组,采用Cox回归评价包括模型风险评分在内的各个临床病理特征对预后的影响;应用R软件绘制生存曲线、构建可视化列线图。应用受试者工作特征(ROC)曲线评估模型预测性能,通过曲线下面积(AUC)评估模型区分度,通过校准曲线和Hosmer-Lemeshow拟合优度检验来评价预测模型的校准度。所有数据的统计分析使用R4.0.2完成,P值多重检验校正采用Benjamini & Hochberg(BH)方法。P<0.05为差异有统计学意义。
2 结果 2.1 HNRNPA2B1在TP53突变肺腺癌中表达的生物信息学分析Kaplan-Meier分析显示,TP53突变影响肺腺癌病人的预后(P=0.033,图 1A),即与非TP53突变肺腺癌病人相比,TP53突变肺腺癌病人总生存期显著缩短;HNRNPA2B1在TP53突变肺腺癌病人中显著高表达(图 1B)。其次,HNRNPA2B1表达水平与肺腺癌预后相关(P=0.031)(图 1C),即与低表达HNRNPA2B1的肺腺癌病人比较,肺腺癌病人HNRNPA2B1表达升高导致其总生存期显著缩短。根据HNRNPA2B1表达分为高表达、低表达两组进行GSEA分析,结果显示HNRNPA2B1表达升高会导致多个通路中某些产物上调,包括气味结合、嗅觉受体活性、调节血管平滑肌细胞增殖、感觉感知(图 1D)。应用Spearman相关性分析,共筛选出522个与HNRNPA2B1表达呈正相关的miRNAs;应用Kaplan-Meier预后分析,筛选出35个在TP53突变肺腺癌病人中表达越高预后越差的miRNAs,同时符合以上两个条件miRNAs共12个(图 1E)。
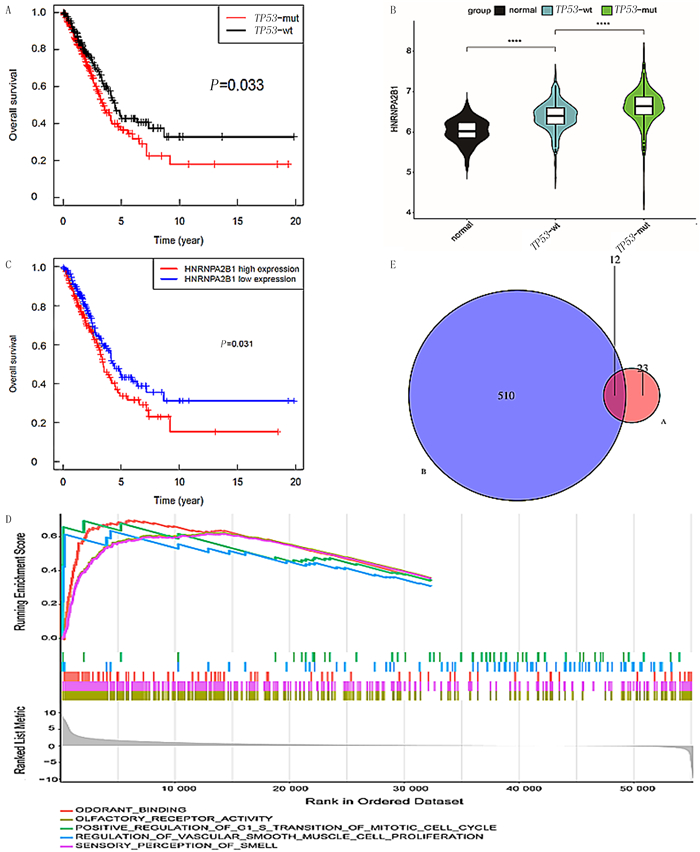 |
| A:与非TP53突变肺腺癌病人相比,TP53突变肺腺癌病人总生存期显著缩短(P=0.033);B:HNRNPA2B1在TP53突变肺腺癌病人中显著高表达;C:与低表达HNRNPA2B1的肺腺癌病人相比,HNRNPA2B1升高导致总生存期显著缩短(P=0.031)。D:GSEA分析显示,HNRNPA2B1高表达会导致多个通路上调;E:Spearman相关分析筛选出522个与HNRNPA2B1表达呈正相关的miRNAs,Kaplan-Meier预后分析选出35个在TP53突变的肺腺癌病人中高表达miRNAs,符合以上两个条件交集miRNAs共12个。****P<0.000 1。 图 1 HNRNPA2B1在TP53突变肺腺癌中表达的生物信息学分析 |
对正常人、TP53-wt组和TP53-mut组中12个HNRNPA2B1潜在调控miRNAs的表达分析显示,相较于正常人及TP53-wt组,TP53-mut组有9个miRNAs(hsa-miR-96、hsa-miR-137、hsa-miR-5001、hsa-miR-940、hsa-miR-616、hsa-miR-3909、hsa-miR-31、hsa-miR-301a、hsa-miR-556)表达水平增高,差异有统计学意义(P<0.05)。见图 2。
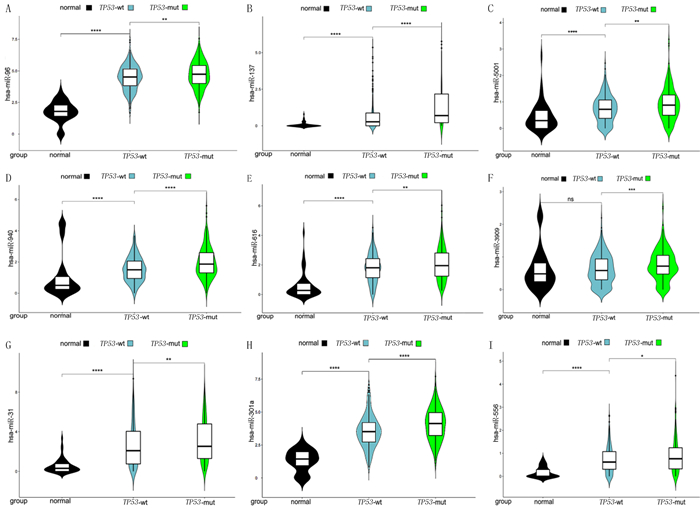 |
| A~I:hsa-miR-96、hsa-miR-137、hsa-miR-5001、hsa-miR-940、hsa-miR-616、hsa-miR-3909、hsa-miR-31、hsa-miR-301a、hsa-miR-556分别在正常组(normal)、TP53-wt组、TP53-mut组中的表达情况。*P<0.05,**P<0.01,***P<0.001,****P<0.000 1。 图 2 TP53突变的肺腺癌中受HNRNPA2B1调控的miRNAs |
基于以上9个miRNAs绘制列线图模型,见图 3A。对模型分析显示,has-miR-940、has-miR-616和HNRNPA2B1表达评分显示为97.5+100.0+57.5=255.0分,与之对应的病人3年生存概率约为76%。训练集和验证集的1、2、3、4年预后预测ROC曲线AUC分别为0.836、0.732、0.716、0.762(图 3B)和0.808、0.829、0.755、0.790(图 3C),说明该模型区分度较好。模型校准曲线见图 3D,Hosmer-Lemeshow检验显示,χ2=9.443,P=0.306,说明该模型有较好校准度。该列线图模型对肺腺癌TP53突变病人的预后具有较好的预测能力。
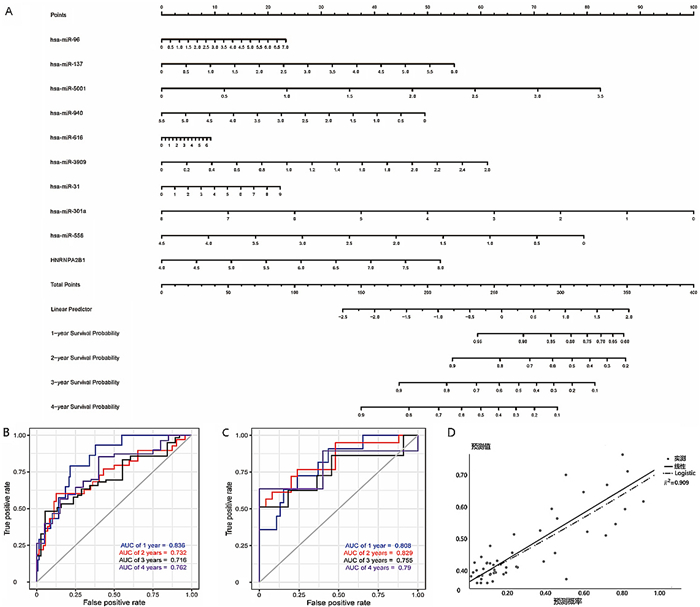 |
| A:训练集9个miRNAs列线图模型评分;B: 训练集9个miRNAs列线图模型计算1、2、3、4年预后预测AUC值分别为0.836、0.732、0.716、0.762;C:验证集的9个miRNAs列线图模型计算1、2、3、4年预后预测AUC值分别为0.808、0.829、0.755、0.790;D:校准曲线。 图 3 TP53突变肺腺癌HNRNPA2B1-miRNAs预后模型建立与评估 |
在NSCLC中,HNRNPA2B1可通过与多种蛋白质相互作用参与致癌过程[16]。然而,目前人们对其与TP53的相互作用是否会影响NSCLC病人的预后,尚知之甚少。猜测TP53突变可能影响了某些潜在受到HNRNPA2B1调控的miRNAs的表达水平,进而影响肺腺癌病人的预后。多篇文献报道HNRNPA2B1通过调控miRNA影响肿瘤发生发展[22]。本文研究通过生物信息学分析筛选出可能潜在受HNRNPA2B1调控的9个miRNAs,分别为miR-96、miR-137、miR-5001、miR-940、miR-616、miR-3909、miR-31、miR-301a和miR-556;它们与HNRNPA2B1在肺腺癌病人中的表达趋势一致,说明以上miRNAs的表达很可能受到HNRNPA2B1调控,随即构建预后模型并用列线图可视化,通过区分度和校准度评估模型准确性。
有研究结果显示,miR-96在NSCLC细胞中可靶向并下调SAMD9的表达,从而减少因顺铂诱导的细胞凋亡[23]。LUO等[24]研究发现,miR-137可通过靶向COX-2调节上皮间质转化(EMT)相关蛋白来抑制NSCLC细胞迁移侵袭。既往研究结果显示,CCMAlnc可通过调节miR-5001-5p与其靶向mRNA之间的相互作用来促进结直肠癌的发生发展[25]。miR-940可以通过靶向NSCLC中Snail 3′-UTR的mRNA来抑制TGF-β诱导的EMT和细胞侵袭[26]。WANG等[27]研究显示,高表达miR-616的NSCLC病人的总生存期和无病生存期显著缩短,且证实了SOX7是miR-616的下游靶点。DAVENPORT等[28]研究显示,与正常肺组织相比,miR-31在肺腺癌、鳞状细胞癌和大细胞神经内分泌癌中过表达,但在小细胞癌中不表达,提示miR-31过表达改变了不同组织学亚型中的细胞信号程序,导致肺癌的表型差异。还有研究发现,miR-301a过表达影响了病人无病生存率,可能作为新的NSCLC预后指标、淋巴结转移的生物标志物以及治疗靶点[29];miR-556-5p下调可诱导NLRP3介导的细胞焦亡,有效提高NSCLC对顺铂的敏感性[30]。
此外,WU等[31]的研究利用环状RNA(circ-ABCB10)与miR-556-3p结合负调控miR-556-3p的表达,上调AK4进而逆转circ-ABCB10低表达,对肺癌顺铂化疗发挥增敏和肿瘤抑制作用。随着测序技术的发展,有研究利用高通量测序技术获得肺癌miRNAs的组织特异性表达谱,利用分子特征区分NSCLC的腺癌和鳞癌亚型,其中包括了表达上调的hsa-miR-556-5p,进一步强调了miRNA信号作为NSCLC分层生物标志物的特殊诊断性能。提示miRNAs在肺癌中具有重要作用。结合本研究结果进一步提示miRNAs在NSCLC中扮演重要角色,与肿瘤的发生发展和病人预后有着极大关系。
综上所述,本研究通过生物信息学分析找到受HNRNPA2B1调控的9个潜在miRNAs,它们的表达显著影响着TP53突变肺腺癌病人的预后情况,可能成为新的肿瘤治疗靶点。此外,本研究构建的预后模型可帮助临床对TP53突变肺腺癌病人的预后判断提供有益的参考。
| [1] |
CHIUL Y, EMERY A, JAIN N, et al. Encoded conformational dynamics of the HIV splice site A3 regulatory locus: implications for differential binding of hnRNP splicing auxiliary factors[J]. Journal of Molecular Biology, 2022, 434(18): 167728. DOI:10.1016/j.jmb.2022.167728 |
| [2] |
TANG J Z, CHEN Z M, WANG Q, et al. hnRNPA2B1 promotes colon cancer progression via the MAPK pathway[J]. Frontiersin Genetics, 2021, 12: 666451. DOI:10.3389/fgene.2021.666451 |
| [3] |
GEUENS T, BOUHY D, TIMMERMAN V. The hnRNP family: insights into their role in health and disease[J]. Human Genetics, 2016, 135(8): 851-867. DOI:10.1007/s00439-016-1683-5 |
| [4] |
ZHANG Y H, HUANG W B, YUAN Y J, et al. Long non-coding RNA H19 promotes colorectal cancer metastasis via binding to hnRNPA2B1[J]. Journal of Experimental & Clinical Cancer Research, 2020, 39(1): 141. DOI:10.3760/cma.j.issn.1001-9030.2020.01.041 |
| [5] |
MAKOWCZENKOK G, JASTRZEBSKIJ P, KIEZUN M, et al. Adaptation of the porcine pituitary transcriptome, spliceosome and editome during early pregnancy[J]. International Journal of Molecular Sciences, 2023, 24(6): 5946. DOI:10.3390/ijms24065946 |
| [6] |
MUSLIMOVI A, TUZHILIN A, TANGT H, et al. Interactions of noncanonical motifs with hnRNP A2 promote activity-dependent RNA transport in neurons[J]. The Journal of Cell Biology, 2014, 205(4): 493-510. DOI:10.1083/jcb.201310045 |
| [7] |
LIU H, LI D X, SUN L N, et al. Interaction of lncRNA MIR100HG with hnRNPA2B1 facilitates m6A-dependent stabilization of TCF7L2 mRNA and colorectal cancer progression[J]. Molecular Cancer, 2022, 21(1): 74. DOI:10.1186/s12943-022-01555-3 |
| [8] |
SHI X, RAN L, LIU Y, et al. Knockdown of hnRNP A2/B1 inhibits cell proliferation, invasion and cell cycle triggering apoptosis in cervical cancer via PI3K/AKT signaling pathway[J]. Oncology Reports, 2018, 39(3): 939-950. |
| [9] |
CHEN T, GUC X, XUEC L, et al. LncRNA-uc002mbe.2 interacting with hnRNPA2B1 mediates AKT deactivation and p21 up-regulation induced by trichostatin in liver cancer cells[J]. Frontiersin Pharmacology, 2017, 8: 669. DOI:10.3389/fphar.2017.00669 |
| [10] |
PETRIB J, PIELLK M, SOUTH WHITTG C, et al. HNRNPA2B1 regulates tamoxifen- and fulvestrant-sensitivity and hallmarks of endocrine resistance in breast cancer cells[J]. Cancer Letters, 2021, 518: 152-168. DOI:10.1016/j.canlet.2021.07.015 |
| [11] |
HU Y, SUNZ H, DENGJ M, et al. Splicing factor hnRNPA2B1 contributes to tumorigenic potential of breast cancer cells through STAT3 and ERK1/2 signaling pathway[J]. Tumour Biology, 2017, 39(3): 1010428317694318. |
| [12] |
YUP F, KANGA R, JINGL J, et al. Long non-coding RNA CACNA1G-AS1 promotes cell migration, invasion and epithelial-mesenchymal transition by HNRNPA2B1 in non-small cell lung cancer[J]. European Reviewfor Medicaland Pharmacolo-gical Sciences, 2018, 22(4): 993-1002. |
| [13] |
CHENX Y, ZHANG J, ZHUJ S. The role of m6A RNA methylation in human cancer[J]. Molecular Cancer, 2019, 18(1): 103. DOI:10.1186/s12943-019-1033-z |
| [14] |
HUNGC Y, WANGY C, CHUANGJ Y, et al. Nm23-H1-stabilized hnRNPA2/B1 promotes internal ribosomal entry site (IRES)-mediated translation of Sp1 in the lung cancer progression[J]. Scientific Reports, 2017, 7(1): 9166. DOI:10.1038/s41598-017-09558-7 |
| [15] |
GAOL B, ZHUX L, SHIJ X, et al. HnRNPA2B1 promotes the proliferation of breast cancer MCF-7 cells via the STAT3 pathway[J]. Journal of Cellular Biochemistry, 2021, 122(3-4): 472-484. DOI:10.1002/jcb.29875 |
| [16] |
CHEN Z Y, CHEN X, LEI T Y, et al. Integrative analysis of NSCLC identifies LINC01234 as an oncogenic lncRNA that interacts with HNRNPA2B1 and regulates miR-106b biogenesis[J]. Molecular Therapy, 2020, 28(6): 1479-1493. DOI:10.1016/j.ymthe.2020.03.010 |
| [17] |
VOKESN I, CHAMBERS E, NGUYEN T, et al. Concurrent TP53 mutations facilitate resistance evolution in EGFR-mutant lung adenocarcinoma[J]. Journal of Thoracic Oncology: Official Publication of the International Association for the Study of Lung Cancer, 2022, 17(6): 779-792. |
| [18] |
LE X N, MOLIFE C, LEUSCH M S, et al. TP53 co-mutation status association with clinical outcomes in patients with EGFR-mutant non-small cell lung cancer[J]. Cancers, 2022, 14(24): 6127. DOI:10.3390/cancers14246127 |
| [19] |
LIU S R, YU J, ZHANG H, et al. TP53 co-mutations in advanced EGFR-mutated non-small cell lung cancer: prognosis and therapeutic strategy for cancer therapy[J]. Frontiers in Oncology, 2022, 12: 860563. DOI:10.3389/fonc.2022.860563 |
| [20] |
ZHAO Z C, WAN J H, GUO M M, et al. Expression and prognostic significance of m6A-related genes in TP53-mutant non-small-cell lung cancer[J]. Journal of Clinical Laboratory Analysis, 2022, 36(1): e24118. DOI:10.1002/jcla.24118 |
| [21] |
LU Y, WANG X Y, GU Q, et al. Heterogeneous nuclear ribonucleoprotein A/B: an emerging group of cancer biomarkers and therapeutic targets[J]. Cell Death Discovery, 2022, 8(1): 337. DOI:10.1038/s41420-022-01129-8 |
| [22] |
JIN C H, CAO J L, CAI Y, et al. A nomogram for predicting the risk of invasive pulmonary adenocarcinoma for patients with solitary peripheral subsolid nodules[J]. The Journal of Thoracic and Cardiovascular Surgery, 2017, 153(2): 462-469.e1. DOI:10.1016/j.jtcvs.2016.10.019 |
| [23] |
WU L, PU X X, WANG Q Z, et al. miR-96 induces cisplatin chemoresistance in non-small cell lung cancer cells by downregulating SAMD9[J]. Oncology Letters, 2016, 11(2): 945-952. DOI:10.3892/ol.2015.4000 |
| [24] |
LUO Y T, HU S W, WANG F, et al. MiR-137 represses migration and cell motility by targeting COX-2 in non-small cell lung cancer[J]. Translational Cancer Research, 2022, 11(10): 3803-3813. DOI:10.21037/tcr-22-2177 |
| [25] |
YAN Y Q, XUAN B Q, GAO Z Y, et al. CCMAlnc promotes the malignance of colorectal cancer by modulating the interaction between miR-5001-5p and its target mRNA[J]. Frontiers in Cell and Developmental Biology, 2020, 8: 566932. DOI:10.3389/fcell.2020.566932 |
| [26] |
JIANG K Q, ZHAO T, SHEN M J, et al. MiR-940 inhibits TGF-β-induced epithelial-mesenchymal transition and cell invasion by targeting Snail in non-small cell lung cancer[J]. Journal of Cancer, 2019, 10(12): 2735-2744. DOI:10.7150/jca.31800 |
| [27] |
WANG D P, CAO Q F, QU M J, et al. Retracted Micro-RNA-616 promotes the growth and metastasis of non-small cell lung cancer by targeting SOX7[J]. Oncology Reports, 2023, 50(4): 186. DOI:10.3892/or.2023.8623 |
| [28] |
DAVENPORTM L, ECHOLSJ B, SILVAA D, et al. MiR-31 displays subtype specificity in lung cancer[J]. Cancer Research, 2021, 81(8): 1942-1953. DOI:10.1158/0008-5472.CAN-20-2769 |
| [29] |
SHIY K, ZANGQ L, LIG X, et al. Increased expression of microRNA-301a in nonsmall-cell lung cancer and its clinical significance[J]. Journal of Cancer Researchand Therapeutics, 2016, 12(2): 693-698. DOI:10.4103/0973-1482.146130 |
| [30] |
SHI F, ZHANG L Q, LIU X, et al. Knock-down of micro-RNA miR-556-5p increases cisplatin-sensitivity in non-small cell lung cancer (NSCLC) via activating NLR family pyrin domain containing 3 (NLRP3)-mediated pyroptotic cell death[J]. Bioengineered, 2021, 12(1): 6332-6342. DOI:10.1080/21655979.2021.1971502 |
| [31] |
WU Z H, GONG Q, YU Y, et al. Knockdown of circ-ABCB10 promotes sensitivity of lung cancer cells to cisplatin via miR-556-3p/AK4 axis[J]. BMC Pulmonary Medicine, 2020, 20(1): 10. DOI:10.1186/s12890-019-1035-z |
 2024, Vol. 60
2024, Vol. 60

