心肌肥大是提高心肌储备能力和心排血量的一种适应性代偿性反应,病理性心肌肥大与许多心脏疾病如高血压、冠心病等具有共同的病理过程,不可逆转,严重危害人类健康[1-2]。目前,心肌肥大的研究涉及蛋白修饰、DNA修饰和RNA修饰等方面,但尚不够全面[3-6],其在转录调控方面的研究还处于起始阶段。染色质开放性测序技术(ATAC-seq)是一种在全基因组范围内研究转录调控的利器[7],已被广泛应用于研究小鼠视觉皮质[8]、心肌梗死[9]、肿瘤[10]以及免疫系统[11]中关键的转录调控因子以及新的调控机制。本研究使用血管紧张素Ⅱ(Ang Ⅱ)构建心肌肥大模型,应用转录组测序(RNA-seq)和ATAC-seq两种高通量测序方法,探讨全基因组内的基因差异表达以及部分特征基因顺式调控元件区域的染色质可及性,从两个不同层次水平上提供心肌肥大发生后的数据,以揭示影响特征基因差异表达的潜在因素,加深对驱动心肌肥大的分子变化的认识,为后期揭示心肌肥大基因差异表达的潜在调控机制以及更深层次信号通路和分子水平方面的研究提供数据支持。现将结果报告如下。
1 材料和方法 1.1 动物分组及处理SPF级8周雄性C57BL/6小鼠12只,购自青岛大任富城畜牧有限公司。小鼠适应性饲养7 d后,随机分为对照组和心肌肥大组,每组6只,分别腹腔注射生理盐水100 μL和AngⅡ(购自EMD Millipore公司,用生理盐水溶解,0.6 mg/(kg·d))100 μL。
1.2 超声心动图检查腹腔注射2周后,对小鼠进行超声检测,检测指标包括室间隔舒张末厚度(IVSd)、左室舒张末期内径(LVIDd)、舒张末左室后壁厚度(LVPWd)、室间隔收缩末厚度(IVSs)、左心室收缩期内径(LVIDs)、收缩末左室后壁厚度(LVPWs)、射血分数(EF)、缩短分数(FS)和心率(HR)等。
1.3 心肌细胞的大小及纤维化程度检测采用免疫组化方法。超声检查完成后,腹腔注射适量的水合氯醛(40 g/L)麻醉,断颈处死小鼠,随后用剪刀剪开小鼠胸腔,取出心脏,使用40 g/L多聚甲醛固定,然后分别行苏木精-伊红(HE)染色和Masson染色检测心肌细胞的大小及纤维化程度。
1.4 RNA-seq检测基因表达取心肌组织,按照Trizol(购自Takara公司)说明书提取总RNA,用NanoDrop ND-1000型分光光度仪进行质量检测,然后送华大基因应用RNA-seq方法检测基因表达情况。
1.5 差异表达基因的筛选与分析方法根据测序所获得的基因表达变化量的log2值进行筛选,最后用DAVID网站进行GO功能分析和KEGG通路分析。
1.6 染色质开放性检测制备细胞核悬液,然后依次进行转座、纯化和DNA文库扩增,最后送华大基因应用ATAC-seq检测染色质开放性。
1.7 实时荧光定量PCR(qPCR)检测采用qPCR检测基因的表达量,使用逆转录试剂盒(购自Takara公司)将RNA逆转录为cDNA,然后应用TB Green Premix Ex Taq Ⅱ试剂盒进行qPCR,并用2-△△Ct评估基因的相对表达量。实验重复3次,取均值。
1.8 统计学分析使用SPSS 20.0软件进行统计学分析。计量资料结果以x±s表示,数据间比较采用t检验。P<0.05表示差异有统计学意义。
2 结果 2.1 两组超声心动图比较与对照组比较,心肌肥大组小鼠的LVIDd、EF和FS均显著下降,LVPWd和LVPWs均显著升高,差异有显著性(t=-5.616~8.750, P < 0.05);两组IVSd、IVSs、LVIDs和HR比较,差异无统计学意义(P>0.05)。见表 1和图 1。表明心肌肥大模型建立成功。
| 表 1 两组小鼠超声心动图检查结果比较(n=6, x±s) |
|
|
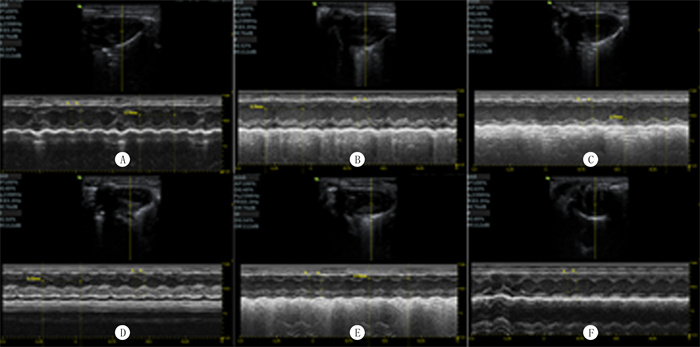 |
| A、B、C:对照组3个重复;D、E、F:心肌肥大组3个重复。 图 1 小鼠心脏的超声心动图检查 |
对照组心肌细胞为椭圆形,排列整齐,细胞核大小均一,心肌纤维排列整齐(图 2A)。心肌肥大组心肌细胞肥大,心肌纤维出现不同程度增生,排列紊乱(图 2B)。
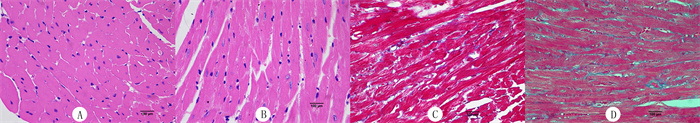 |
| A: 对照组HE染色;B:心肌肥大组HE染色;C:对照组Masson染色;D: 心肌肥大组Masson染色。400倍。 图 2 两组心肌免疫组化染色检查 |
Masson染色心肌细胞为红色, 胶原为蓝色。对照组心肌细胞间隙没有胶原增生(图 2C)。心肌肥大组心肌细胞间质纤维化并有胶原堆积(图 2D)。进一步证明心肌肥大模型建立成功。
2.4 基因差异表达情况应用生物信息学工具对基因进行GO生物学进程分析和KEGG通路分析(图 3A、B、C)。结果表明,GO生物学进程分析主要富集到炎症反应、免疫反应、血管生成、心肌收缩和钙稳态等生物学功能;KEGG通路分析主要富集到PI3K-Akt通路、心肌肥大、钙离子信号通路、Rap1通路、cAMP通路、Ras通路、Foxo通路、AMPK通路、cGMP-PKG通路和MAPK通路等。筛选出27个与心肌肥大相关的基因(图 3D),其中8个基因表达下调,分别为α肌球蛋白重链(myh6)、Foxp1、Sirt3、Sirt2、Ppargc1a、Ppargc1b、心肌肌浆网Ca2+-ATP酶2a(Atp2a2)、Trim63;19个表达上调,分别为心房钠尿肽(ANP)、脑钠肽(BNP)、α骨骼肌肌动蛋白(Acta1)、β-肌球蛋白重链(myh7)、Col3a1、Col1a1、CYP1B1、Fn1、Il18、CD36、Timp1、Tgfbr1、PDE1C、PDE1A、Sirt1、Sirt4、Pfkp、CDK1和Fbln2。筛选出23个与心肌疾病相关的基因(图 3E),其中13个表达下调,分别为DSP、Timp3、Sgcb、Hfe、Ldb3、Fktn、Apln、Fkrp、Prkce、Sgca、Pdgfa、Prkaa2、Gja1;10个表达上调,分别为Mapk8、Rtn4、Dcn、Myot、Ptk2b、Ppbp、Xdh、Tnfrsf1b、Ctnnb1、Cxadr 。
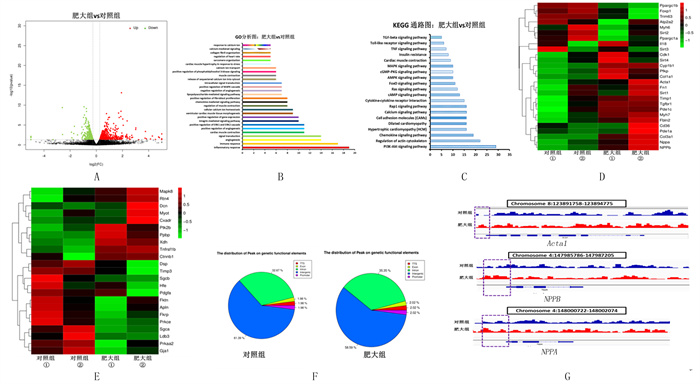 |
| A:差异表达基因的火山图;B:差异表达基因GO功能分析图;C: 差异表达基因KEGG通路分析图;D:肥大相关基因的聚类热图;E:心肌病相关基因的聚类热图;F:peak在不同顺式调控区域分布情况;G:可视化部分肥大marker基因的ATAC-seq信号。 图 3 基因表达图谱的差异性分析 |
开放结构主要位于基因间区域,其次是内含子区域;基因的外显子、启动子和转录起始位点区域的peak占比相同(图 3F)。心肌肥大后,基因间区域的peak在总体中占比有所下降,而内含子、外显子、启动子和转录起始位点区域的peak占比有所增加。通过IGV软件可视化分析部分肥大基因相应区域,结果表明心肌肥大后,ATAC-seq信号在NPPA、NPPB和Acta1 转录起始位点附近区域有所增强;此外,在肥大发生之前这些基因就存在一定的开放性(图 3G)。
2.6 两组marker基因比较qPCR检测结果显示,心肌肥大组marker基因ANP、BNP、β-MHC和Acta1表达较对照组显著升高,差异有显著性(t=4.685~7.124,P < 0.05)。见图 4。说明心肌发生了明显肥大。
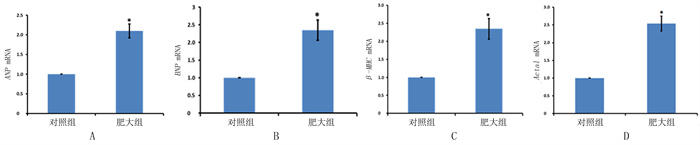 |
| 图 4 部分肥大marker基因的qPCR结果 |
心肌肥大是心脏为了应对内、外部生物学应力刺激而做出的应激反应,严重危害人类健康[1]。心肌肥大主要特点是心室壁变厚,心肌细胞体积变大,并伴有心肌功能障碍和纤维化。本研究超声检测和免疫组化结果均说明本研究制备的心肌肥大模型特征与其一致。
本研究差异表达基因的GO功能和KEGG通路分析得到的心肌肥大相关通路与NAKAMURA等[12]研究结果相同。心肌肥大发生后,与心肌肥大相关的基因随之发生差异表达,其中,marker基因ANP、BNP、Acta1、myh7等的表达升高,而myh6的表达下降,进一步证明心肌肥大模型构建成功。在心肌肥大相关的基因中,Col3a1、Col1a1、Fn1、Il18、Timp1和Tgfbr1与心肌纤维化和胶原生成相关,其表达升高说明心肌发生纤维化,这与相关研究结果一致[3-6, 13-16]。另有研究表明,PDE1既参与心脏病理性重塑和纤维化,又与钙调蛋白相互作用,通过PKG通路调控心肌肥大[3]。Fbln2与TGFβ共同调控TAK1发生磷酸化,继而激活P38/JNK、CaN-NFAT和NF-kB信号通路,促进心肌肥大[17]。Foxp1参与心脏重塑、心肌纤维化、成肌纤维细胞形成、细胞外基质蛋白生成和心肌肥大[18],这些研究所涉及的心肌肥大相关基因在本研究中也呈现出相同的表达结果。此外,在心肌肥大相关基因中,一些基因涉及心肌功能、代谢功能和心律失常,如Sirt1、Sirt2、Sirt3和Sirt4等与线粒体能量代谢密切相关。有研究表明,心肌肥大后去乙酰化酶的持久性失活会导致线粒体蛋白高度乙酰化和能量代谢紊乱,Sirt3引发线粒体相关蛋白高度乙酰化,导致心肌肥大和纤维化[12]。而CYP1B1、Ppargc1a和Ppargc1与心肌肥大的代谢重编程相关,CD36和Atp2a2能导致心律失常和心功能障碍,Trim63、Pfkp和CDK1参与调控心肌肥大[19-23]。
另外,本文研究在差异表达的基因中还获得一些与心肌疾病相关的基因,其中Tnfrsf1b和Prkce与代谢疾病相关[21],Myot和Ldb3与肌原纤维性心肌病相关[24],Fktn和Fkrp与肌营养不良相关的心肌病相关[25],Ppbp是潜在的冠心病标志物[26],Hfe参与血色素沉着症诱发的心肌病[27],Prkaa2在进展性心肌病中发挥作用[21],Timp3参与铁超负荷介导的心肌病[28]。除此以外,本研究在差异表达的基因中,还获得一些涉及心脏功能的基因,其中Ptk2b、Pdgfa、Dcn和Apln调控心脏纤维化[29-32],CXADR和DSP介导心律失常[33-34],而Mapk8和Ct- nnb1分别是肾素-血管紧张素通路和Wnt通路的核心参与者[21, 35],Rtn4调节血管生成[36],Gja1和Xdh与线粒体功能相关[28, 37],Sgca和Sgcb是肌糖复合物形成的必要成分[21]。基因差异表达分析表明,AngⅡ在诱导心肌肥大的过程中,除了能导致心肌肥大以外,还可能导致与心肌疾病和心肌功能相关基因发生差异表达。
本文研究分析ATAC-seq信号在不同顺式调控元件区域上的分布,结果显示,心肌肥大组中启动子和转录起始位点区域的peak所占比例较对照组有所增加,这与其参与调控基因表达的功能密切相关。通过对心肌肥大marker基因相应区域分析显示,心肌肥大后,在NPPA、NPPB和Acta1转录起始位点附近区域的染色质开放性有所增强,这可能为它们表达升高的原因。
综上所述,本文应用RNA-seq和ATAC-seq两种高通量测序技术,研究心肌肥大后基因表达以及特征基因的染色质开放性。结果显示,心肌肥大后,与心肌肥大和心肌疾病相关的基因表达具有明显的差异,心肌肥大特征基因(如ANP、BNP、Acta1 )在染色质开放水平上明显提高,揭示了导致心肌肥大特征基因表达升高的潜在因素。本文在两个不同层次水平上检测了心肌肥大发生后的数据,为后期更深层次地研究心肌肥大相关基因转录调控机制,以及更详细地了解驱动心肌细胞肥大的分子变化提供了数据支持,更为开发逆转心肌肥大病理性重塑的治疗方法提供了参考。
| [1] |
WU M P, ZHANG Y S, XU X B, et al. Vinpocetine atte-nuates pathological cardiac remodeling by inhibiting cardiac hypertrophy and fibrosis[J]. Cardiovascular Drugs and Therapy, 2017, 31(2): 157-166. DOI:10.1007/s10557-017-6719-0 |
| [2] |
KOENTGES C, PEPIN M E, MVSSE C, et al. Gene expression analysis to identify mechanisms underlying heart failure susceptibility in mice and humans[J]. Basic Research in Car-diology, 2018, 113(1): 8. DOI:10.1007/s00395-017-0666-6 |
| [3] |
ZHANG S Q, LU Y, JIANG C Y. Inhibition of histone de-methylase JMJD1C attenuates cardiac hypertrophy and fibrosis induced by angiotensin Ⅱ[J]. Journal of Receptor and Signal Transduction Research, 2020, 40(4): 339-347. DOI:10.1080/10799893.2020.1734819 |
| [4] |
FIX C, BINGHAM K, CARVER W. Effects of interleukin-18 on cardiac fibroblast function and gene expression[J]. Cytokine, 2011, 53(1): 19-28. DOI:10.1016/j.cyto.2010.10.002 |
| [5] |
WO Y, GUO J, LI P H, et al. Long non-coding RNA CHRF facilitates cardiac hypertrophy through regulating Akt3 via miR-93[J]. Cardiovascular Pathology, 2018, 35: 29-36. DOI:10.1016/j.carpath.2018.04.003 |
| [6] |
WANG K, LONG B, LIU F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223[J]. European Heart Journal, 2016, 37(33): 2602-2611. DOI:10.1093/eurheartj/ehv713 |
| [7] |
SUN Y Y, MIAO N, SUN T. Detect accessible chromatin using ATAC-sequencing, from principle to applications[J]. Hereditas, 2019, 156: 29. DOI:10.1186/s41065-019-0105-9 |
| [8] |
GRAY L T, YAO Z Z, NGUYEN T N, et al. Layer-specific chromatin accessibility landscapes reveal regulatory networks in adult mouse visual cortex[J]. eLife, 2017, 6: e21883. DOI:10.7554/eLife.21883 |
| [9] |
VAN DUIJVENBODEN K, DE BAKKER D E M, MAN J C K, et al. Conserved NPPB + border zone switches from MEF2-to AP-1-driven gene program[J]. Circulation, 2019, 140(10): 864-879. DOI:10.1161/CIRCULATIONAHA.118.038944 |
| [10] |
DAVIE K, JACOBS J, ATKINS M, et al. Discovery of transcription factors and regulatory regions driving in vivo tumor development by ATAC-seq and FAIRE-seq open chromatin profiling[J]. PLoS Genetics, 2015, 11(2): e1004994. DOI:10.1371/journal.pgen.1004994 |
| [11] |
YOSHIDA H, LAREAU C A, RAMIREZ R N, et al. The Cis-regulatory atlas of the mouse immune system[J]. Cell, 2019, 176(4): 897-912.e20. DOI:10.1016/j.cell.2018.12.036 |
| [12] |
NAKAMURA M, SADOSHIMA J. Mechanisms of physiolo-gical and pathological cardiac hypertrophy[J]. Nature Reviews Cardiology, 2018, 15(7): 387-407. DOI:10.1038/s41569-018-0007-y |
| [13] |
YAN X Y, ZHAO R, FENG X R, et al. Sialyltransferase7A promotes angiotensin Ⅱ-induced cardiomyocyte hypertrophy via HIF-1α-TAK1 signalling pathway[J]. Cardiovascular Research, 2020, 116(1): 114-126. DOI:10.1093/cvr/cvz064 |
| [14] |
VIGIL-GARCIA M, DEMKES C J, EDING J E C, et al. Gene expression profiling of hypertrophic cardiomyocytes identifies new players in pathological remodelling[J]. Cardiovascular Research, 2021, 117(6): 1532-1545. DOI:10.1093/cvr/cvaa233 |
| [15] |
YAN X Y, ZHAO R, FENG X R, et al. Sialyltransferase7A promotes angiotensin Ⅱ-induced cardiomyocyte hypertrophy via HIF-1α-TAK1 signalling pathway[J]. Cardiovascular Research, 2020, 116(1): 114-126. DOI:10.1093/cvr/cvz064 |
| [16] |
HONG Y, CAO H M, WANG Q, et al. MiR-22 may suppress fibrogenesis by targeting TGFβR I in cardiac fibroblasts[J]. Cellular Physiology and Biochemistry: International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology, 2016, 40(6): 1345-1353. DOI:10.1159/000453187 |
| [17] |
MIAO R J, LU Y, HE X, et al. Ubiquitin-specific protease 19 blunts pathological cardiac hypertrophy via inhibition of the TAK1-dependent pathway[J]. Journal of Cellular and Molecular Medicine, 2020, 24(18): 10946-10957. DOI:10.1111/jcmm.15724 |
| [18] |
LIU J, ZHUANG T, PI J J, et al. Endothelial forkhead box transcription factor P1 regulates pathological cardiac remodeling through transforming growth factor-β1-endothelin-1 signal pathway[J]. Circulation, 2019, 140(8): 665-680. DOI:10.1161/CIRCULATIONAHA.119.039767 |
| [19] |
TSE M M, ABOUTABL M E, ALTHURWI H N, et al. Cy-tochrome P450 epoxygenase metabolite, 14, 15-EET, protects against isoproterenol-induced cellular hypertrophy in H9c2 rat cell line[J]. Vascular Pharmacology, 2013, 58(5/6): 363-373. |
| [20] |
ZHANG Y J, BAO M W, DAI M Y, et al. Cardiospecific CD36 suppression by Lentivirus-mediated RNA interference prevents cardiac hypertrophy and systolic dysfunction in high-fat-diet induced obese mice[J]. Cardiovascular Diabetology, 2015, 14: 69. DOI:10.1186/s12933-015-0234-z |
| [21] |
AUERBACH S S, THOMAS R, SHAH R, et al. Comparative phenotypic assessment of cardiac pathology, physiology, and gene expression in C3H/HeJ, C57BL/6J, and B6C3F1/J mice[J]. Toxicologic Pathology, 2010, 38(6): 923-942. DOI:10.1177/0192623310382864 |
| [22] |
WILLIS M S, WADOSKY K M, RODRÍGUEZ J E, et al. Muscle ring finger 1 and muscle ring finger 2 are necessary but functionally redundant during developmental cardiac growth and regulate E2F1-mediated gene expression in vivo[J]. Cell Biochemistry and Function, 2014, 32(1): 39-50. DOI:10.1002/cbf.2969 |
| [23] |
MAEJIMA Y, USUI S, ZHAI P Y, et al. Muscle-specific RING finger 1 negatively regulates pathological cardiac hypertrophy through downregulation of calcineurin A[J]. Circulation Heart Failure, 2014, 7(3): 479-490. DOI:10.1161/CIRCHEARTFAILURE.113.000713 |
| [24] |
AVILA-SMIRNOW D, GUENEAU L, BATONNET-PICHON S, et al. Cardiac arrhythmia and late-onset muscle weakness caused by a myofibrillar myopathy with unusual histopathological features due to a novel missense mutation in FLNC[J]. Revue Neurologique, 2016, 172(10): 594-606. DOI:10.1016/j.neurol.2016.07.017 |
| [25] |
UJIHARA Y, KANAGAWA M, MOHRI S, et al. Elimination of fukutin reveals cellular and molecular pathomechanisms in muscular dystrophy-associated heart failure[J]. Nature Communications, 2019, 10(1): 5754. DOI:10.1038/s41467-019-13623-2 |
| [26] |
MANEERAT Y, PRASONGSUKARN K, BENJATHUMMARAK S, et al. PPBP and DEFA1/DEFA3 genes in hyperlipidaemia as feasible synergistic inflammatory biomarkers for coronary heart disease[J]. Lipids in Health and Disease, 2017, 16(1): 80. DOI:10.1186/s12944-017-0471-0 |
| [27] |
DJEMAI H, THOMASSON R, TRZASKUS Y, et al. A mouse model of cardiomyopathy induced by mutations in the hemochromatosis HFE gene[J]. The Canadian Journal of Cardiology, 2017, 33(7): 904-910. DOI:10.1016/j.cjca.2017.03.006 |
| [28] |
FAN D, KASSIRI Z. Biology of tissue inhibitor of metalloproteinase 3 (TIMP3), and its therapeutic implications in cardiovascular pathology[J]. Frontiers in Physiology, 2020, 11: 661. DOI:10.3389/fphys.2020.00661 |
| [29] |
BALASUBRAMANIAN S, QUINONES L, KASIGANESAN H, et al. β3 integrin in cardiac fibroblast is critical for extracellular matrix accumulation during pressure overload hypertrophy in mouse[J]. PLoS One, 2012, 7(9): e45076. DOI:10.1371/journal.pone.0045076 |
| [30] |
GALLINI R, LINDBLOM P, BONDJERS C, et al. PDGF-A and PDGF-B induces cardiac fibrosis in transgenic mice[J]. Experimental Cell Research, 2016, 349(2): 282-290. DOI:10.1016/j.yexcr.2016.10.022 |
| [31] |
YAN W, WANG P H, ZHAO C X, et al. Decorin gene deli-very inhibits cardiac fibrosis in spontaneously hypertensive rats by modulation of transforming growth factor-beta/Smad and p38 mitogen-activated protein kinase signaling pathways[J]. Human Gene Therapy, 2009, 20(10): 1190-1200. DOI:10.1089/hum.2008.204 |
| [32] |
SATO T, KADOWAKI A, SUZUKI T, et al. Loss of apelin augments angiotensin Ⅱ-induced cardiac dysfunction and pa-thological remodeling[J]. International Journal of Molecular Sciences, 2019, 20(2): E239. DOI:10.3390/ijms20020239 |
| [33] |
MARSMAN R F, BEZZINA C R, FREIBERG F, et al. Coxsackie and adenovirus receptor is a modifier of cardiac conduction and arrhythmia vulnerability in the setting of myocardial ischemia[J]. Journal of the American College of Cardiology, 2014, 63(6): 549-559. DOI:10.1016/j.jacc.2013.10.062 |
| [34] |
DUBASH A D, KAM C Y, AGUADO B A, et al. Plakophi-lin-2 loss promotes TGF-β1/p38 MAPK-dependent fibrotic gene expression in cardiomyocytes[J]. The Journal of Cell Biology, 2016, 212(4): 425-438. DOI:10.1083/jcb.201507018 |
| [35] |
ZHENG Q L, CHEN P, XU Z Q, et al. Expression and redistribution of β-catenin in the cardiac myocytes of left ventricle of spontaneously hypertensive rat[J]. Journal of Molecular Histology, 2013, 44(5): 565-573. DOI:10.1007/s10735-013-9507-6 |
| [36] |
BULLARD T A, PROTACK T L, AGUILAR F, et al. Identification of Nogo as a novel indicator of heart failure[J]. Phy-siological Genomics, 2008, 32(2): 182-189. DOI:10.1152/physiolgenomics.00200.2007 |
| [37] |
FU Y L, TAO L, PENG F H, et al. GJA1-20k attenuates Ang Ⅱ-induced pathological cardiac hypertrophy by regulating gap junction formation and mitochondrial function[J]. Acta Pharmacologica Sinica, 2021, 42(4): 536-549. DOI:10.1038/s41401-020-0459-6 |
 2022, Vol. 58
2022, Vol. 58

